One of the things I say when teaching about fingerprints and similarity searches is that there is no best fingerprint for every use case and that it’s often useful to do similarity searches using different types of fingerprints and combine the results. If you use fingerprints that use quite different features to determine similarity (e.g. Morgan3 and atom pairs), this is a good way to take advantage of the differences between the fingerprints.
The goal of this post is to explore (and try to quantitate) the differences between the hit sets returned by doing similarity searches with different fingerprint types.
For this post I will use the 59788 compounds tested in PubChem Bioassay ID 373 . This is one of the PubChem screening assays, so the the compounds are taken from a screening deck and should be expected to be reasonably diverse with some small SAR clusters. You can directly access and download the compound structures here .
One thing that is worth keeping in mind is that these results almost certainly consider similarity values which are down close to and probably in the region of similarities observed between random compounds . It may be worth refining the analysis in order to only consider similarities which are more significant, but that’s for a possible future post.
from rdkit import Chemfrom rdkit.Chem import rdFingerprintGeneratorfrom rdkit.Chem import Drawfrom rdkit.Chem.Draw import IPythonConsolefrom rdkit import DataStructsimport numpy as npfrom matplotlib import pyplot as plt'tableau-colorblind10' )% matplotlib inline% load_ext sqlimport rdkitprint (rdkit.__version__)
import gzipwith gzip.open ('../data/Pubchem_AID373_compounds.sdf.gz' ,'rb' ) as inf:with Chem.ForwardSDMolSupplier(inf) as suppl:= [x for x in suppl if x is not None ]len (dbmols)
= Chem.MolFromSmiles('CCOC1=C(C=C(C=C1)C2=NOC(=N2)C3=CC=NC=C3)OCC' ) # https://pubchem.ncbi.nlm.nih.gov/compound/666359
= ['mfp2' ,rdFingerprintGenerator.GetMorganGenerator(radius= 2 )),'ffp2' ,rdFingerprintGenerator.GetMorganGenerator(radius= 2 ,= rdFingerprintGenerator.GetMorganAtomInvGen())),'mfp3' ,rdFingerprintGenerator.GetMorganGenerator(radius= 3 )),'ffp3' ,rdFingerprintGenerator.GetMorganGenerator(radius= 3 ,= rdFingerprintGenerator.GetMorganAtomInvGen())),'mfp1' ,rdFingerprintGenerator.GetMorganGenerator(radius= 1 )),'ffp1' ,rdFingerprintGenerator.GetMorganGenerator(radius= 1 ,= rdFingerprintGenerator.GetMorganAtomInvGen())),'tt' ,rdFingerprintGenerator.GetTopologicalTorsionGenerator()),'ap' ,rdFingerprintGenerator.GetAtomPairGenerator()),'rdk5' ,rdFingerprintGenerator.GetRDKitFPGenerator(maxPath= 5 )),'rdk7' ,rdFingerprintGenerator.GetRDKitFPGenerator(maxPath= 7 )),= {}for nm,fpg in runs:print (nm)= fpg.GetFingerprints(dbmols,numThreads= 8 )= DataStructs.BulkTanimotoSimilarity(fpg.GetFingerprint(qry),fps)f' { nm} -b' ] = sims= fpg.GetCountFingerprints(dbmols,numThreads= 8 )= DataStructs.BulkTanimotoSimilarity(fpg.GetCountFingerprint(qry),list (fps))f' { nm} -c' ] = sims
mfp2
ffp2
mfp3
ffp3
mfp1
ffp1
tt
ap
rdk5
rdk7
from rdkit.Chem.Pharm2D import Gobbi_Pharm2D,Generatedef Gobbi2D_bits(mol,fpLen= 2048 ):= DataStructs.ExplicitBitVect(fpLen)for bit in Generate.Gen2DFingerprint(mol,Gobbi_Pharm2D.factory).GetOnBits():# the bits are not hashed, so we need to do so before we fold them: hash ((bit,))% fpLen)return res
from rdkit.Chem import rdMolDescriptorsfrom rdkit.Avalon import pyAvalonTools= ['gobbi2d' ,Gobbi2D_bits),'avalon' ,pyAvalonTools.GetAvalonFP),'avalon-c' ,pyAvalonTools.GetAvalonCountFP),'pattern' ,Chem.PatternFingerprint),for nm,func in func_runs:print (nm)= [func(m) for m in dbmols]= DataStructs.BulkTanimotoSimilarity(func(qry),fps)= sims
gobbi2d
avalon
avalon-c
pattern
Accumulate the overlaps between the hit sets:
from collections import defaultdict= list (accum.keys())= defaultdict(dict )= [10 ,100 ,1000 ]for i,nmi in enumerate (nms):= [x for s,x in sorted ([(s,x) for x,s in enumerate (accum[nmi])],reverse= True )]for j in range (i):= nms[j]= [x for s,x in sorted ([(s,x) for x,s in enumerate (accum[nmj])],reverse= True )]for topN in topNs:= set (topi[:topN]).intersection(topj[:topN])= dict (ovls)
Here’s the full table of concordances between the fingerprints in the top 10, 100, and 1000 results:
from IPython.display import HTML= sorted (nms)= "<table>" = "" .join(f"<th> { nm} </th>" for nm in snms)+= f"<tr><td></td> { ths} </tr> \n " = dict (ovls)for i,nm in enumerate (snms):= f"<th> { nm} </th>" = []for j in range (len (snms)):if i== j:'<td></td>' )continue = snms[i]= snms[j]if (nmi,nmj) not in ovls:= nmj,nmif"<td> { len (ovls[nmi,nmj][10 ])} <br /> { len (ovls[nmi,nmj][100 ])} <br /> { len (ovls[nmi,nmj][1000 ])} </td>" )= "" .join(tds)+= tds+= f"<tr> { row} </tr> \n " += "</table>"
ap-b
ap-c
avalon
avalon-c
ffp1-b
ffp1-c
ffp2-b
ffp2-c
ffp3-b
ffp3-c
gobbi2d
mfp1-b
mfp1-c
mfp2-b
mfp2-c
mfp3-b
mfp3-c
pattern
rdk5-b
rdk5-c
rdk7-b
rdk7-c
tt-b
tt-c
ap-b
9
7
7
6
8
7
7
7
6
3
7
8
7
7
7
7
5
6
6
7
6
7
7
ap-c
9
6
6
6
8
7
7
7
7
3
6
7
6
6
6
6
5
6
6
6
7
7
7
avalon
7
6
9
7
8
8
9
8
8
5
8
9
9
9
8
9
6
7
8
8
8
8
8
avalon-c
7
6
9
7
8
8
9
8
8
5
8
9
9
9
8
9
6
7
8
8
8
8
8
ffp1-b
6
6
7
7
7
9
8
9
8
6
9
7
8
8
8
8
5
7
8
8
7
8
8
ffp1-c
8
8
8
8
7
8
9
8
8
5
7
9
8
8
7
8
6
8
8
7
8
9
9
ffp2-b
7
7
8
8
9
8
9
10
9
6
9
8
9
9
9
9
5
8
8
9
8
9
9
ffp2-c
7
7
9
9
8
9
9
9
9
6
8
9
9
9
8
9
6
8
9
8
9
9
9
ffp3-b
7
7
8
8
9
8
10
9
9
6
9
8
9
9
9
9
5
8
8
9
8
9
9
ffp3-c
6
7
8
8
8
8
9
9
9
6
8
8
9
9
8
9
6
8
8
8
9
9
9
gobbi2d
3
3
5
5
6
5
6
6
6
6
5
5
6
6
5
6
3
7
7
5
5
6
6
mfp1-b
7
6
8
8
9
7
9
8
9
8
5
8
9
9
9
9
5
7
7
9
7
8
8
mfp1-c
8
7
9
9
7
9
8
9
8
8
5
8
9
9
8
9
6
7
8
8
8
8
8
mfp2-b
7
6
9
9
8
8
9
9
9
9
6
9
9
10
9
10
6
8
8
9
8
9
9
mfp2-c
7
6
9
9
8
8
9
9
9
9
6
9
9
10
9
10
6
8
8
9
8
9
9
mfp3-b
7
6
8
8
8
7
9
8
9
8
5
9
8
9
9
9
5
7
7
9
7
8
8
mfp3-c
7
6
9
9
8
8
9
9
9
9
6
9
9
10
10
9
6
8
8
9
8
9
9
pattern
5
5
6
6
5
6
5
6
5
6
3
5
6
6
6
5
6
5
6
5
6
6
6
rdk5-b
6
6
7
7
7
8
8
8
8
8
7
7
7
8
8
7
8
5
8
7
7
9
9
rdk5-c
6
6
8
8
8
8
8
9
8
8
7
7
8
8
8
7
8
6
8
7
8
8
8
rdk7-b
7
6
8
8
8
7
9
8
9
8
5
9
8
9
9
9
9
5
7
7
7
8
8
rdk7-c
6
7
8
8
7
8
8
9
8
9
5
7
8
8
8
7
8
6
7
8
7
8
8
tt-b
7
7
8
8
8
9
9
9
9
9
6
8
8
9
9
8
9
6
9
8
8
8
10
tt-c
7
7
8
8
8
9
9
9
9
9
6
8
8
9
9
8
9
6
9
8
8
8
10
Look at the highest overlaps (most similar results) for each FP:
= sorted (nms)= dict (ovls)for i in range (len (snms)):print ('-' * 70 )print (snms[i])for cnt in (10 ,100 ,1000 ):= []for j in range (len (snms)):if i== j:continue = snms[i]= snms[j]if (nmi,nmj) not in ovls:= nmj,nmilen (ovls[nmi,nmj][cnt]),j))= sorted (row,reverse= True )= []for j in range (3 ):f' { snms[row[j][1 ]]} ( { row[j][0 ]} )' )print (f' \t { cnt: 5d} ' ,f' { nbrs[0 ]:13s} ' ,f' { nbrs[1 ]:13s} ' ,f' { nbrs[2 ]:13s} ' ,)
----------------------------------------------------------------------
ap-b
10 ap-c(9) mfp1-c(8) ffp1-c(8)
100 ap-c(77) gobbi2d(51) rdk7-b(47)
1000 ap-c(724) ffp1-c(383) mfp1-c(380)
----------------------------------------------------------------------
ap-c
10 ap-b(9) ffp1-c(8) tt-c(7)
100 ap-b(77) rdk5-b(52) rdk5-c(51)
1000 ap-b(724) ffp1-c(445) mfp1-c(424)
----------------------------------------------------------------------
avalon
10 mfp3-c(9) mfp2-c(9) mfp2-b(9)
100 rdk5-b(83) rdk5-c(74) rdk7-b(73)
1000 rdk5-b(625) avalon-c(588) mfp1-b(552)
----------------------------------------------------------------------
avalon-c
10 mfp3-c(9) mfp2-c(9) mfp2-b(9)
100 avalon(70) rdk5-c(69) rdk7-c(68)
1000 avalon(588) pattern(532) rdk5-b(494)
----------------------------------------------------------------------
ffp1-b
10 mfp1-b(9) ffp3-b(9) ffp2-b(9)
100 mfp1-b(82) ffp2-b(68) mfp2-b(67)
1000 mfp1-b(759) ffp2-b(671) mfp2-b(662)
----------------------------------------------------------------------
ffp1-c
10 tt-c(9) tt-b(9) mfp1-c(9)
100 mfp1-c(80) ffp2-c(70) mfp2-c(68)
1000 ffp2-c(722) mfp1-c(674) ffp3-c(633)
----------------------------------------------------------------------
ffp2-b
10 ffp3-b(10) tt-c(9) tt-b(9)
100 mfp2-b(84) mfp3-b(82) ffp3-b(82)
1000 ffp3-b(804) ffp2-c(682) ffp1-b(671)
----------------------------------------------------------------------
ffp2-c
10 tt-c(9) tt-b(9) rdk7-c(9)
100 ffp3-c(90) mfp2-c(81) mfp3-c(80)
1000 ffp3-c(848) ffp1-c(722) ffp2-b(682)
----------------------------------------------------------------------
ffp3-b
10 ffp2-b(10) tt-c(9) tt-b(9)
100 mfp3-b(86) mfp3-c(82) ffp2-b(82)
1000 ffp2-b(804) ffp3-c(715) ffp2-c(669)
----------------------------------------------------------------------
ffp3-c
10 tt-c(9) tt-b(9) rdk7-c(9)
100 ffp2-c(90) mfp3-c(82) ffp3-b(81)
1000 ffp2-c(848) ffp3-b(715) ffp2-b(649)
----------------------------------------------------------------------
gobbi2d
10 rdk5-c(7) rdk5-b(7) tt-c(6)
100 mfp2-b(55) ap-b(51) mfp3-b(49)
1000 rdk7-c(371) rdk7-b(367) rdk5-c(367)
----------------------------------------------------------------------
mfp1-b
10 rdk7-b(9) mfp3-c(9) mfp3-b(9)
100 ffp1-b(82) mfp2-b(68) mfp1-c(66)
1000 ffp1-b(759) mfp2-b(729) mfp1-c(645)
----------------------------------------------------------------------
mfp1-c
10 mfp3-c(9) mfp2-c(9) mfp2-b(9)
100 ffp1-c(80) mfp2-c(75) mfp3-c(69)
1000 mfp2-c(729) ffp1-c(674) mfp1-b(645)
----------------------------------------------------------------------
mfp2-b
10 mfp3-c(10) mfp2-c(10) tt-c(9)
100 ffp2-b(84) mfp3-b(83) mfp2-c(81)
1000 mfp3-b(780) mfp1-b(729) mfp2-c(694)
----------------------------------------------------------------------
mfp2-c
10 mfp3-c(10) mfp2-b(10) tt-c(9)
100 mfp3-c(89) mfp3-b(84) mfp2-b(81)
1000 mfp3-c(844) mfp1-c(729) mfp2-b(694)
----------------------------------------------------------------------
mfp3-b
10 rdk7-b(9) mfp3-c(9) mfp2-c(9)
100 mfp3-c(89) ffp3-b(86) mfp2-c(84)
1000 mfp2-b(780) mfp3-c(735) mfp2-c(679)
----------------------------------------------------------------------
mfp3-c
10 mfp2-c(10) mfp2-b(10) tt-c(9)
100 mfp3-b(89) mfp2-c(89) ffp3-c(82)
1000 mfp2-c(844) mfp3-b(735) ffp3-c(645)
----------------------------------------------------------------------
pattern
10 tt-c(6) tt-b(6) rdk7-c(6)
100 avalon-c(62) mfp1-b(57) avalon(56)
1000 avalon-c(532) avalon(518) rdk5-b(513)
----------------------------------------------------------------------
rdk5-b
10 tt-c(9) tt-b(9) rdk5-c(8)
100 rdk7-b(84) rdk5-c(83) avalon(83)
1000 rdk5-c(736) rdk7-b(705) rdk7-c(663)
----------------------------------------------------------------------
rdk5-c
10 ffp2-c(9) tt-c(8) tt-b(8)
100 rdk7-c(86) rdk7-b(85) rdk5-b(83)
1000 rdk7-c(812) rdk5-b(736) rdk7-b(701)
----------------------------------------------------------------------
rdk7-b
10 mfp3-c(9) mfp3-b(9) mfp2-c(9)
100 rdk7-c(85) rdk5-c(85) rdk5-b(84)
1000 rdk7-c(776) rdk5-b(705) rdk5-c(701)
----------------------------------------------------------------------
rdk7-c
10 ffp3-c(9) ffp2-c(9) tt-c(8)
100 rdk5-c(86) rdk7-b(85) rdk5-b(73)
1000 rdk5-c(812) rdk7-b(776) rdk5-b(663)
----------------------------------------------------------------------
tt-b
10 tt-c(10) rdk5-b(9) mfp3-c(9)
100 tt-c(87) mfp2-b(74) ffp2-b(70)
1000 tt-c(853) ffp2-b(595) ffp2-c(578)
----------------------------------------------------------------------
tt-c
10 tt-b(10) rdk5-b(9) mfp3-c(9)
100 tt-b(87) mfp2-b(75) ffp2-b(74)
1000 tt-b(853) ffp2-b(611) ffp2-c(603)
Do some statistics
Those were just results for a single query molecule. Let’s try and get more robust results by picking 500 molecules and looking at statistics for the overlaps:
import random0xbad5eed )= list (range (len (dbmols)))= 500 = order[:500 ]= {}for nm,fpg in runs:print (nm)= list (fpg.GetFingerprints(dbmols,numThreads= 8 ))= []for i in order:= fps[i]= [(sim,j) for j,sim in enumerate (DataStructs.BulkTanimotoSimilarity(qry,fps))]# remove the self similarity del sims[i]= sorted (sims,reverse= True )[:1000 ]f' { nm} -b' ] = taccum= list (fpg.GetCountFingerprints(dbmols,numThreads= 8 ))= []for i in order:= fps[i]= [(sim,j) for j,sim in enumerate (DataStructs.BulkTanimotoSimilarity(qry,fps))]# remove the self similarity del sims[i]= sorted (sims,reverse= True )[:1000 ]f' { nm} -c' ] = taccumfor nm,func in func_runs:print (nm)= [func(m) for m in dbmols]= []for i in order:= fps[i]= [(sim,j) for j,sim in enumerate (DataStructs.BulkTanimotoSimilarity(qry,fps))]# remove the self similarity del sims[i]= sorted (sims,reverse= True )[:1000 ]= taccum
mfp2
ffp2
mfp3
ffp3
mfp1
ffp1
tt
ap
rdk5
rdk7
gobbi2d
avalon
avalon-c
pattern
import pickleimport gzipwith gzip.open ('./results/sim_overlaps.pkl.gz' ,'wb+' ) as outf:
Accumulate the overlaps:
from collections import defaultdict= list (accum.keys())= defaultdict(dict )= [10 ,100 ,1000 ]for i,nmi in enumerate (nms):= len (accum[nmi])for j in range (i):= nms[j]for topN in topNs:= []for run in range (nruns):= [x for s,x in accum[nmi][run]]= [x for s,x in accum[nmj][run]]for topN in topNs:len (set (topi[:topN]).intersection(topj[:topN])))= dict (ovls)
Find the most similar fingerprint types for each of the fingerprints, this time using the mean overlap across the 500 hit sets:
= sorted (nms)for i in range (len (snms)):print ('-' * 70 )print (snms[i])for cnt in (10 ,100 ,1000 ):= []for j in range (len (snms)):if i== j:continue = snms[i]= snms[j]if (nmi,nmj) not in ovls:= nmj,nmi= sorted (row,reverse= True )= []for j in range (3 ):f' { snms[row[j][1 ]]:8s} ( { row[j][0 ]:5.1f} )' )print (f' \t { cnt: 5d} ' ,f' { nbrs[0 ]:17s} ' ,f' { nbrs[1 ]:17s} ' ,f' { nbrs[2 ]:16s} ' ,)
----------------------------------------------------------------------
ap-b
10 ap-c ( 8.1) ffp1-c ( 5.0) mfp1-c ( 5.0)
100 ap-c ( 74.9) ffp1-c ( 40.7) mfp1-c ( 39.7)
1000 ap-c (732.9) ffp1-c (395.4) mfp1-c (370.4)
----------------------------------------------------------------------
ap-c
10 ap-b ( 8.1) ffp2-c ( 5.1) ffp1-c ( 5.1)
100 ap-b ( 74.9) ffp1-c ( 43.6) mfp1-c ( 41.9)
1000 ap-b (732.9) ffp1-c (432.4) mfp1-c (402.0)
----------------------------------------------------------------------
avalon
10 avalon-c( 5.6) rdk7-b ( 5.4) rdk5-b ( 5.3)
100 avalon-c( 49.7) rdk5-b ( 49.3) rdk7-b ( 48.1)
1000 avalon-c(452.4) rdk5-b (403.7) rdk5-c (370.2)
----------------------------------------------------------------------
avalon-c
10 avalon ( 5.6) rdk5-c ( 5.4) rdk7-c ( 5.1)
100 avalon ( 49.7) rdk5-c ( 45.9) rdk7-c ( 43.2)
1000 avalon (452.4) rdk5-c (378.0) rdk7-c (358.5)
----------------------------------------------------------------------
ffp1-b
10 mfp1-b ( 7.2) ffp2-b ( 6.7) mfp2-b ( 6.2)
100 mfp1-b ( 66.2) ffp2-b ( 64.0) mfp2-b ( 56.8)
1000 ffp2-b (645.2) mfp1-b (622.2) mfp2-b (545.6)
----------------------------------------------------------------------
ffp1-c
10 mfp1-c ( 7.6) ffp2-c ( 7.0) mfp2-c ( 6.5)
100 mfp1-c ( 68.0) ffp2-c ( 66.5) mfp2-c ( 58.3)
1000 ffp2-c (698.4) mfp1-c (652.9) ffp3-c (610.5)
----------------------------------------------------------------------
ffp2-b
10 ffp3-b ( 8.2) mfp2-b ( 7.9) mfp3-b ( 7.5)
100 ffp3-b ( 80.2) mfp2-b ( 74.1) mfp3-b ( 70.4)
1000 ffp3-b (797.6) mfp2-b (679.3) mfp3-b (651.9)
----------------------------------------------------------------------
ffp2-c
10 ffp3-c ( 8.3) mfp2-c ( 8.1) mfp3-c ( 7.5)
100 ffp3-c ( 82.2) mfp2-c ( 74.4) mfp3-c ( 70.8)
1000 ffp3-c (840.0) mfp2-c (700.4) ffp1-c (698.4)
----------------------------------------------------------------------
ffp3-b
10 ffp2-b ( 8.2) mfp3-b ( 8.1) mfp2-b ( 7.5)
100 ffp2-b ( 80.2) mfp3-b ( 74.7) ffp3-c ( 69.5)
1000 ffp2-b (797.6) mfp3-b (662.3) ffp3-c (660.8)
----------------------------------------------------------------------
ffp3-c
10 ffp2-c ( 8.3) mfp3-c ( 8.2) mfp2-c ( 7.6)
100 ffp2-c ( 82.2) mfp3-c ( 75.4) mfp2-c ( 70.6)
1000 ffp2-c (840.0) mfp3-c (692.7) mfp2-c (674.0)
----------------------------------------------------------------------
gobbi2d
10 ap-c ( 3.9) ap-b ( 3.8) mfp3-c ( 3.6)
100 ap-c ( 27.7) ap-b ( 27.6) mfp2-c ( 27.0)
1000 ap-b (253.4) ap-c (251.7) mfp2-c (218.5)
----------------------------------------------------------------------
mfp1-b
10 ffp1-b ( 7.2) mfp2-b ( 7.0) ffp2-b ( 6.3)
100 mfp2-b ( 67.2) ffp1-b ( 66.2) ffp2-b ( 57.9)
1000 mfp2-b (681.8) ffp1-b (622.2) mfp3-b (581.7)
----------------------------------------------------------------------
mfp1-c
10 ffp1-c ( 7.6) mfp2-c ( 7.2) ffp2-c ( 6.5)
100 mfp2-c ( 69.8) ffp1-c ( 68.0) ffp2-c ( 60.0)
1000 mfp2-c (727.6) ffp1-c (652.9) mfp3-c (644.3)
----------------------------------------------------------------------
mfp2-b
10 mfp3-b ( 8.3) ffp2-b ( 7.9) ffp3-b ( 7.5)
100 mfp3-b ( 81.1) ffp2-b ( 74.1) ffp3-b ( 69.1)
1000 mfp3-b (807.9) mfp1-b (681.8) ffp2-b (679.3)
----------------------------------------------------------------------
mfp2-c
10 mfp3-c ( 8.3) ffp2-c ( 8.1) ffp3-c ( 7.6)
100 mfp3-c ( 83.4) ffp2-c ( 74.4) ffp3-c ( 70.6)
1000 mfp3-c (847.8) mfp1-c (727.6) ffp2-c (700.4)
----------------------------------------------------------------------
mfp3-b
10 mfp2-b ( 8.3) ffp3-b ( 8.1) ffp2-b ( 7.5)
100 mfp2-b ( 81.1) ffp3-b ( 74.7) mfp3-c ( 71.4)
1000 mfp2-b (807.9) mfp3-c (678.4) ffp3-b (662.3)
----------------------------------------------------------------------
mfp3-c
10 mfp2-c ( 8.3) ffp3-c ( 8.2) ffp2-c ( 7.5)
100 mfp2-c ( 83.4) ffp3-c ( 75.4) mfp3-b ( 71.4)
1000 mfp2-c (847.8) ffp3-c (692.7) ffp2-c (678.8)
----------------------------------------------------------------------
pattern
10 avalon-c( 4.8) avalon ( 4.3) rdk5-c ( 4.2)
100 avalon-c( 42.3) avalon ( 39.9) rdk5-c ( 38.9)
1000 avalon-c(356.0) rdk5-c (345.9) rdk5-b (340.6)
----------------------------------------------------------------------
rdk5-b
10 rdk7-b ( 7.3) rdk5-c ( 6.4) rdk7-c ( 6.1)
100 rdk7-b ( 68.4) rdk5-c ( 64.8) rdk7-c ( 59.3)
1000 rdk5-c (651.3) rdk7-c (545.5) rdk7-b (535.0)
----------------------------------------------------------------------
rdk5-c
10 rdk7-c ( 7.5) rdk5-b ( 6.4) rdk7-b ( 6.1)
100 rdk7-c ( 74.9) rdk5-b ( 64.8) rdk7-b ( 58.2)
1000 rdk7-c (688.7) rdk5-b (651.3) rdk7-b (457.5)
----------------------------------------------------------------------
rdk7-b
10 rdk5-b ( 7.3) rdk7-c ( 7.0) rdk5-c ( 6.1)
100 rdk5-b ( 68.4) rdk7-c ( 63.8) rdk5-c ( 58.2)
1000 rdk7-c (538.0) rdk5-b (535.0) rdk5-c (457.5)
----------------------------------------------------------------------
rdk7-c
10 rdk5-c ( 7.5) rdk7-b ( 7.0) rdk5-b ( 6.1)
100 rdk5-c ( 74.9) rdk7-b ( 63.8) rdk5-b ( 59.3)
1000 rdk5-c (688.7) rdk5-b (545.5) rdk7-b (538.0)
----------------------------------------------------------------------
tt-b
10 tt-c ( 8.6) ffp3-c ( 6.2) mfp3-c ( 6.1)
100 tt-c ( 83.4) ffp2-b ( 54.9) mfp2-b ( 53.5)
1000 tt-c (824.8) ffp2-b (481.9) ffp2-c (468.5)
----------------------------------------------------------------------
tt-c
10 tt-b ( 8.6) ffp3-c ( 6.4) ffp2-c ( 6.3)
100 tt-b ( 83.4) ffp2-c ( 54.9) ffp3-c ( 54.6)
1000 tt-b (824.8) ffp2-c (494.2) ffp2-b (486.5)
Look at histograms of the overlap sizes for a few different fingerprint pairs
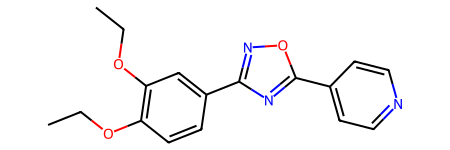
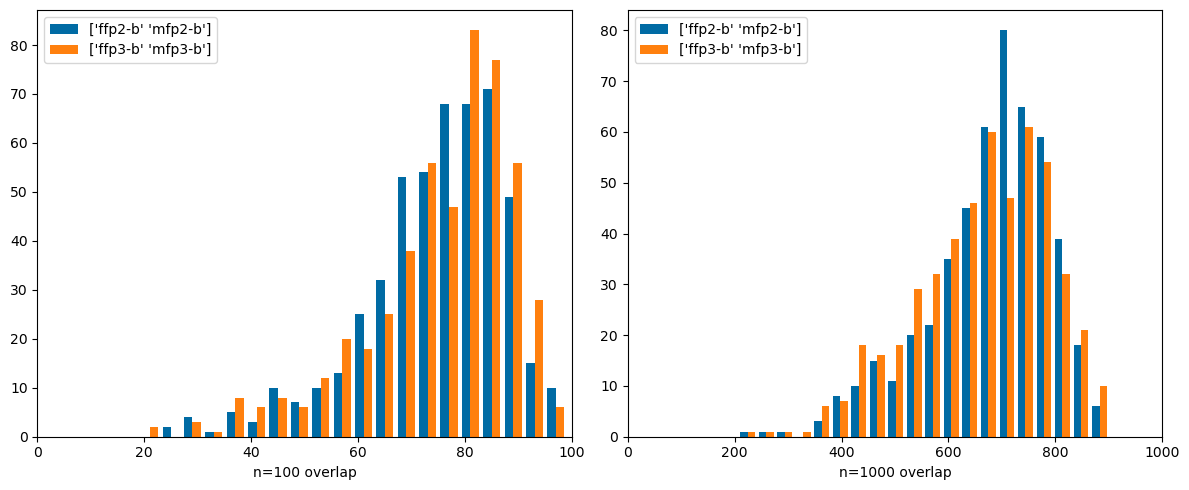
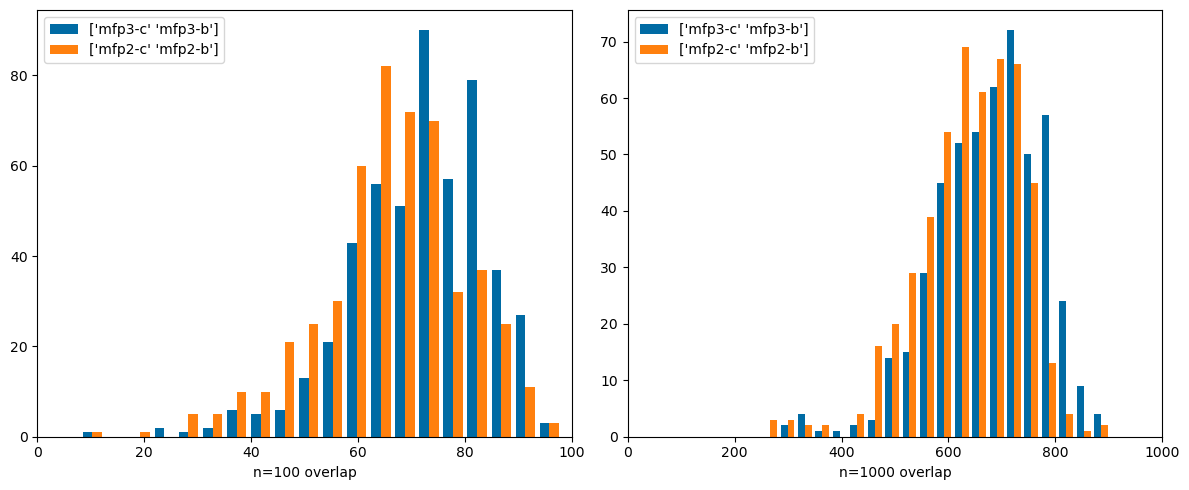
def compare(prs,ovls= ovls):= list (prs)for i,pr in enumerate (prs):if pr not in ovls:= pr[1 ],pr[0 ]= pr= (12 ,5 ))for i,n in enumerate ((100 ,1000 )):1 ,2 ,i+ 1 )for pr in prs],label= prs,bins= 20 ); 0 ,n)f'n= { n} overlap' )#plt.title(pr)
Start with FeatureMorgan and Morgan , where there is a reasonably large amount of overlap:
'ffp2-b' ,'mfp2-b' ),('ffp3-b' ,'mfp3-b' )))
Now compare count-based and bit-based Morgan fingerprints. I expected the overlaps here to be higher:
'mfp3-c' ,'mfp3-b' ),('mfp2-c' ,'mfp2-b' ),))
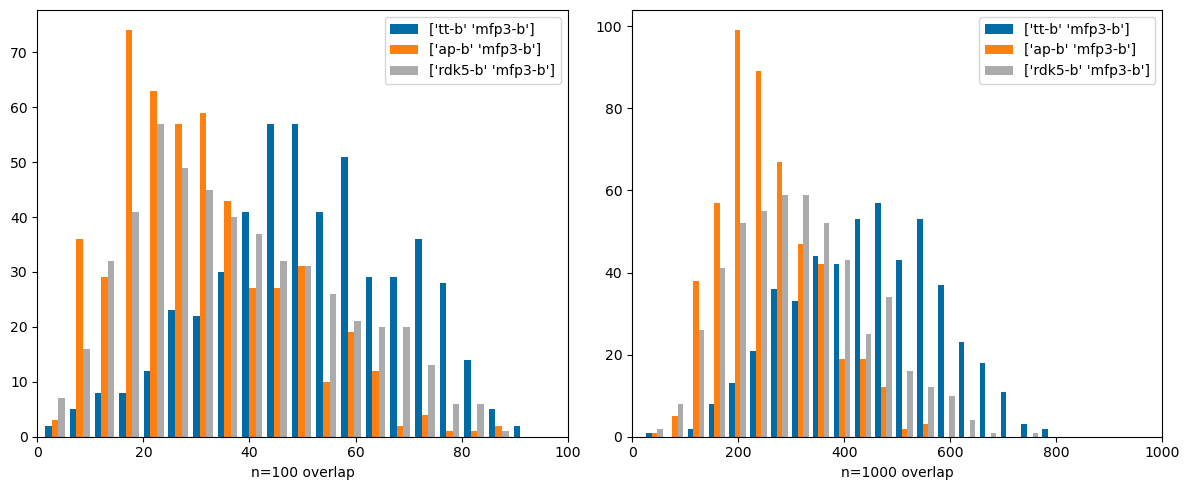
Comparing very different types of fingerprints: count-based Morgan3 and both Topological Torsions and Atom Pairs (these both use count simulation . Here there are significant differences. These are fingerprints that it would be interesting to use together.
'tt-b' ,'mfp3-c' ),('ap-b' ,'mfp3-c' )))
Same thing with bit-baseed Morgan3 and RDK5 . Again, These are nicely complementary fingerprints:
'tt-b' ,'mfp3-b' ),('ap-b' ,'mfp3-b' ),('rdk5-b' ,'mfp3-b' )))
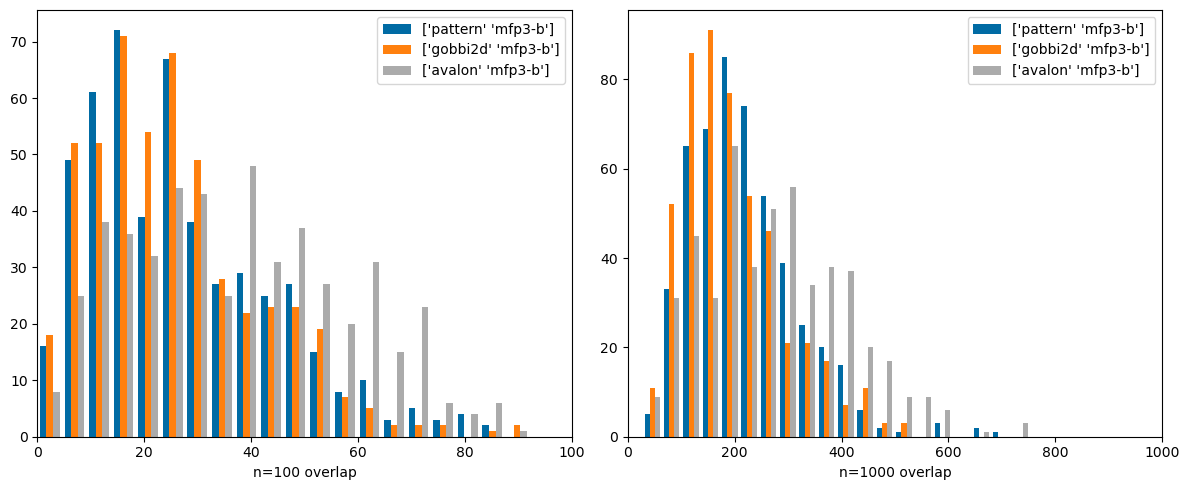
Finally compare Morgan3 with the pattern fingerprint (normally used for substructure screening, not similarity search), Gobbi2D (a 2D pharmacophore FP) and the Avalon FP. These are also nicely different from each other:
'pattern' ,'mfp3-b' ),('gobbi2d' ,'mfp3-b' ),('avalon' ,'mfp3-b' )))