from rdkit import Chem
from rdkit.Chem import Draw
from rdkit.Chem.Draw import IPythonConsole
from rdkit.Chem import PandasTools
PandasTools.RenderImagesInAllDataFrames(True)Using the ACS1996 drawing style in PandasTools
This is a short one showing how to improve the legibility of small structure drawings in Pandas DataFrames displayed in the notebook.
Start by reading in an SDF:
df = PandasTools.LoadSDF('../data/chembl26_very_active.sdf.gz')
df.head()[04:36:24] Warning: ambiguous stereochemistry - zero final chiral volume - at atom 50 ignored
[04:36:25] Warning: ambiguous stereochemistry - overlapping neighbors - at atom 7 ignored
[04:36:25] Warning: ambiguous stereochemistry - overlapping neighbors - at atom 9 ignored
[04:36:25] Warning: ambiguous stereochemistry - overlapping neighbors - at atom 9 ignored| compound_chembl_id | assay_chembl_id | target_chembl_id | pref_name | standard_relation | standard_value | standard_units | standard_type | ID | ROMol | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CHEMBL86971 | CHEMBL690412 | CHEMBL243 | Human immunodeficiency virus type 1 protease | = | 0.8 | nM | Ki | ||
| 1 | CHEMBL6710 | CHEMBL621508 | CHEMBL324 | Serotonin 2c (5-HT2c) receptor | = | 0.26 | nM | Ki | ||
| 2 | CHEMBL314218 | CHEMBL677606 | CHEMBL248 | Leukocyte elastase | = | 0.03 | nM | Ki | ||
| 3 | CHEMBL438897 | CHEMBL877834 | CHEMBL1855 | Gonadotropin-releasing hormone receptor | = | 0.24 | nM | Ki | ||
| 4 | CHEMBL15928 | CHEMBL616937 | CHEMBL1983 | Serotonin 1d (5-HT1d) receptor | = | 0.7 | nM | Ki |
Move the molecule to the first column
cols = list(df.columns)
cols.insert(0,cols.pop(-1))
df = df[cols]
df.head()| ROMol | compound_chembl_id | assay_chembl_id | target_chembl_id | pref_name | standard_relation | standard_value | standard_units | standard_type | ID | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CHEMBL86971 | CHEMBL690412 | CHEMBL243 | Human immunodeficiency virus type 1 protease | = | 0.8 | nM | Ki | ||
| 1 | CHEMBL6710 | CHEMBL621508 | CHEMBL324 | Serotonin 2c (5-HT2c) receptor | = | 0.26 | nM | Ki | ||
| 2 | CHEMBL314218 | CHEMBL677606 | CHEMBL248 | Leukocyte elastase | = | 0.03 | nM | Ki | ||
| 3 | CHEMBL438897 | CHEMBL877834 | CHEMBL1855 | Gonadotropin-releasing hormone receptor | = | 0.24 | nM | Ki | ||
| 4 | CHEMBL15928 | CHEMBL616937 | CHEMBL1983 | Serotonin 1d (5-HT1d) receptor | = | 0.7 | nM | Ki |
The default drawing style doesn’t look great when the molecule images are small like this; the ACS1996 style really works a lot better.
We can use the ACS996 style in the DataFrame by setting the drawing options for PandasTools. We do this by creating a new drawing options instance, setting that to ACS1996 mode, and then setting the drawOptions variable in the PandasTools module.
dopts = Draw.MolDrawOptions()
Draw.SetACS1996Mode(dopts,Draw.MeanBondLength(df.ROMol[0]))
PandasTools.drawOptions = doptsThe new options are now used when we show the DataFrame:
df.head()| ROMol | compound_chembl_id | assay_chembl_id | target_chembl_id | pref_name | standard_relation | standard_value | standard_units | standard_type | ID | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CHEMBL86971 | CHEMBL690412 | CHEMBL243 | Human immunodeficiency virus type 1 protease | = | 0.8 | nM | Ki | ||
| 1 | CHEMBL6710 | CHEMBL621508 | CHEMBL324 | Serotonin 2c (5-HT2c) receptor | = | 0.26 | nM | Ki | ||
| 2 | CHEMBL314218 | CHEMBL677606 | CHEMBL248 | Leukocyte elastase | = | 0.03 | nM | Ki | ||
| 3 | CHEMBL438897 | CHEMBL877834 | CHEMBL1855 | Gonadotropin-releasing hormone receptor | = | 0.24 | nM | Ki | ||
| 4 | CHEMBL15928 | CHEMBL616937 | CHEMBL1983 | Serotonin 1d (5-HT1d) receptor | = | 0.7 | nM | Ki |
The ACS1996 style will also be used when we do things like generate an XLSX file:
PandasTools.SaveXlsxFromFrame(df.head(100),'../data/output.xlsx')As an aside: we can also set ACS1996 mode for the rendering in the notebook;
Draw.SetACS1996Mode(IPythonConsole.drawOptions,Draw.MeanBondLength(df.ROMol[0]))
df.ROMol[0]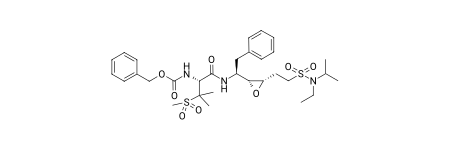
That style is also used in Draw.MolsToGridImage():
Draw.MolsToGridImage(df.ROMol[:8],molsPerRow=4)