from rdkit import Chem
from rdkit.Chem import Draw
from rdkit.Chem.Draw import IPythonConsole
from rdkit.Chem import rdChemReactions
import rdkit
print(rdkit.__version__)2021.09.2November 26, 2021
A while ago there was a question on Twitter about highlighting the bonds which changed in a reaction. I put together a quick bit of example code to answer that question and made a note to do a blog post on the topic. I’m finally getting around to doing that blog post.
Fair warning: This one is heavy on code and light on words. :-)
from rdkit import Chem
from rdkit.Chem import Draw
from rdkit.Chem.Draw import IPythonConsole
from rdkit.Chem import rdChemReactions
import rdkit
print(rdkit.__version__)2021.09.2Here’s something similar to the reaction from the question:
rxn1 = rdChemReactions.ReactionFromRxnBlock('''$RXN
Mrv2102 111820212128
2 1
$MOL
Mrv2102 11182121282D
13 13 0 0 0 0 999 V2000
-7.5723 2.6505 0.0000 C 0 0 0 0 0 0 0 0 0 1 0 0
-6.8579 2.2380 0.0000 O 0 0 0 0 0 0 0 0 0 2 0 0
-6.8580 1.4130 0.0000 C 0 0 0 0 0 0 0 0 0 3 0 0
-6.1435 1.0004 0.0000 O 0 0 0 0 0 0 0 0 0 4 0 0
-7.5725 1.0005 0.0000 C 0 0 0 0 0 0 0 0 0 5 0 0
-7.5725 0.1755 0.0000 N 0 0 0 0 0 0 0 0 0 6 0 0
-8.2869 -0.2369 0.0000 C 0 0 0 0 0 0 0 0 0 7 0 0
-8.2870 -1.0620 0.0000 C 0 0 0 0 0 0 0 0 0 8 0 0
-9.0015 -1.4745 0.0000 C 0 0 0 0 0 0 0 0 0 9 0 0
-9.0015 -2.2995 0.0000 C 0 0 0 0 0 0 0 0 0 10 0 0
-8.2870 -2.7120 0.0000 C 0 0 0 0 0 0 0 0 0 11 0 0
-7.5726 -2.2995 0.0000 C 0 0 0 0 0 0 0 0 0 12 0 0
-7.5726 -1.4745 0.0000 C 0 0 0 0 0 0 0 0 0 13 0 0
1 2 1 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
3 5 1 0 0 0 0
5 6 1 0 0 0 0
6 7 2 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
8 13 2 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
M END
$MOL
Mrv2102 11182121282D
12 11 0 0 0 0 999 V2000
-3.7934 0.7703 0.0000 C 0 0 0 0 0 0 0 0 0 14 0 0
-3.0790 1.1828 0.0000 C 0 0 0 0 0 0 0 0 0 15 0 0
-2.3645 0.7703 0.0000 C 0 0 0 0 0 0 0 0 0 16 0 0
-3.7934 -0.0547 0.0000 C 0 0 0 0 0 0 0 0 0 17 0 0
-4.5078 -0.4672 0.0000 O 0 0 0 0 0 0 0 0 0 18 0 0
-3.0789 -0.4671 0.0000 O 0 0 0 0 0 0 0 0 0 19 0 0
-1.6500 1.1828 0.0000 O 0 0 0 0 0 0 0 0 0 20 0 0
-2.3645 -0.0547 0.0000 O 0 0 0 0 0 0 0 0 0 21 0 0
-3.0788 -1.2922 0.0000 C 0 0 0 0 0 0 0 0 0 22 0 0
-1.6500 -0.4672 0.0000 C 0 0 0 0 0 0 0 0 0 23 0 0
-2.3644 -1.7046 0.0000 C 0 0 0 0 0 0 0 0 0 24 0 0
-1.6500 -1.2922 0.0000 C 0 0 0 0 0 0 0 0 0 25 0 0
1 2 2 0 0 0 0
1 4 1 0 0 0 0
2 3 1 0 0 0 0
3 7 2 0 0 0 0
3 8 1 0 0 0 0
4 5 2 0 0 0 0
4 6 1 0 0 0 0
6 9 1 0 0 0 0
8 10 1 0 0 0 0
9 11 1 0 0 0 0
10 12 1 0 0 0 0
M END
$MOL
Mrv2102 11182121282D
25 26 0 0 0 0 999 V2000
5.1328 0.9532 0.0000 C 0 0 0 0 0 0 0 0 0 5 0 0
5.8002 0.4683 0.0000 N 0 0 0 0 0 0 0 0 0 6 0 0
5.5453 -0.3163 0.0000 C 0 0 0 0 0 0 0 0 0 7 0 0
4.7203 -0.3163 0.0000 C 0 0 0 0 0 0 0 0 0 14 0 0
4.4654 0.4683 0.0000 C 0 0 0 0 0 0 0 0 0 15 0 0
5.1328 1.7782 0.0000 C 0 0 0 0 0 0 0 0 0 3 0 0
3.6807 0.7232 0.0000 C 0 0 0 0 0 0 0 0 0 16 0 0
4.2354 -0.9838 0.0000 C 0 0 0 0 0 0 0 0 0 17 0 0
6.0302 -0.9838 0.0000 C 0 0 0 0 0 0 0 0 0 8 0 0
6.8507 -0.8975 0.0000 C 0 0 0 0 0 0 0 0 0 9 0 0
7.3356 -1.5650 0.0000 C 0 0 0 0 0 0 0 0 0 10 0 0
7.0001 -2.3187 0.0000 C 0 0 0 0 0 0 0 0 0 11 0 0
6.1796 -2.4049 0.0000 C 0 0 0 0 0 0 0 0 0 12 0 0
5.6947 -1.7375 0.0000 C 0 0 0 0 0 0 0 0 0 13 0 0
3.4149 -0.8975 0.0000 O 0 0 0 0 0 0 0 0 0 18 0 0
4.5709 -1.7375 0.0000 O 0 0 0 0 0 0 0 0 0 19 0 0
4.0860 -2.4049 0.0000 C 0 0 0 0 0 0 0 0 0 22 0 0
3.2655 -2.3187 0.0000 C 0 0 0 0 0 0 0 0 0 24 0 0
3.5092 1.5302 0.0000 O 0 0 0 0 0 0 0 0 0 20 0 0
3.0676 0.1712 0.0000 O 0 0 0 0 0 0 0 0 0 21 0 0
2.2830 0.4261 0.0000 C 0 0 0 0 0 0 0 0 0 23 0 0
1.6699 -0.1259 0.0000 C 0 0 0 0 0 0 0 0 0 25 0 0
5.8473 2.1907 0.0000 O 0 0 0 0 0 0 0 0 0 4 0 0
4.4183 2.1907 0.0000 O 0 0 0 0 0 0 0 0 0 2 0 0
4.4183 3.0157 0.0000 C 0 0 0 0 0 0 0 0 0 1 0 0
1 2 1 0 0 0 0
2 3 1 0 0 0 0
3 4 1 0 0 0 0
4 5 1 0 0 0 0
1 5 1 0 0 0 0
1 6 1 0 0 0 0
5 7 1 0 0 0 0
4 8 1 0 0 0 0
3 9 1 0 0 0 0
10 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
9 10 1 0 0 0 0
9 14 2 0 0 0 0
8 15 2 0 0 0 0
8 16 1 0 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
7 19 2 0 0 0 0
7 20 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 0 0 0 0
6 23 2 0 0 0 0
6 24 1 0 0 0 0
24 25 1 0 0 0 0
M END
''')Let’s take a look at the reaction:
We can ask the reaction to tell us which atoms in the reactants are modified in the reaction:
The information about which atoms react is enough to figure out which bonds change, but we have to do some additional work for this:
from collections import namedtuple
AtomInfo = namedtuple('AtomInfo',('mapnum','reactant','reactantAtom','product','productAtom'))
def map_reacting_atoms_to_products(rxn,reactingAtoms):
''' figures out which atoms in the products each mapped atom in the reactants maps to '''
res = []
for ridx,reacting in enumerate(reactingAtoms):
reactant = rxn.GetReactantTemplate(ridx)
for raidx in reacting:
mapnum = reactant.GetAtomWithIdx(raidx).GetAtomMapNum()
foundit=False
for pidx,product in enumerate(rxn.GetProducts()):
for paidx,patom in enumerate(product.GetAtoms()):
if patom.GetAtomMapNum()==mapnum:
res.append(AtomInfo(mapnum,ridx,raidx,pidx,paidx))
foundit = True
break
if foundit:
break
return res
def get_mapped_neighbors(atom):
''' test all mapped neighbors of a mapped atom'''
res = {}
amap = atom.GetAtomMapNum()
if not amap:
return res
for nbr in atom.GetNeighbors():
nmap = nbr.GetAtomMapNum()
if nmap:
if amap>nmap:
res[(nmap,amap)] = (atom.GetIdx(),nbr.GetIdx())
else:
res[(amap,nmap)] = (nbr.GetIdx(),atom.GetIdx())
return res
BondInfo = namedtuple('BondInfo',('product','productAtoms','productBond','status'))
def find_modifications_in_products(rxn):
''' returns a 2-tuple with the modified atoms and bonds from the reaction '''
reactingAtoms = rxn.GetReactingAtoms()
amap = map_reacting_atoms_to_products(rxn,reactingAtoms)
res = []
seen = set()
# this is all driven from the list of reacting atoms:
for _,ridx,raidx,pidx,paidx in amap:
reactant = rxn.GetReactantTemplate(ridx)
ratom = reactant.GetAtomWithIdx(raidx)
product = rxn.GetProductTemplate(pidx)
patom = product.GetAtomWithIdx(paidx)
rnbrs = get_mapped_neighbors(ratom)
pnbrs = get_mapped_neighbors(patom)
for tpl in pnbrs:
pbond = product.GetBondBetweenAtoms(*pnbrs[tpl])
if (pidx,pbond.GetIdx()) in seen:
continue
seen.add((pidx,pbond.GetIdx()))
if not tpl in rnbrs:
# new bond in product
res.append(BondInfo(pidx,pnbrs[tpl],pbond.GetIdx(),'New'))
else:
# present in both reactants and products, check to see if it changed
rbond = reactant.GetBondBetweenAtoms(*rnbrs[tpl])
if rbond.GetBondType()!=pbond.GetBondType():
res.append(BondInfo(pidx,pnbrs[tpl],pbond.GetIdx(),'Changed'))
return amap,resLet’s look at what that function returns for our reaction:
[AtomInfo(mapnum=5, reactant=0, reactantAtom=4, product=0, productAtom=0), AtomInfo(mapnum=6, reactant=0, reactantAtom=5, product=0, productAtom=1), AtomInfo(mapnum=7, reactant=0, reactantAtom=6, product=0, productAtom=2), AtomInfo(mapnum=14, reactant=1, reactantAtom=0, product=0, productAtom=3), AtomInfo(mapnum=15, reactant=1, reactantAtom=1, product=0, productAtom=4)]
[BondInfo(product=0, productAtoms=(4, 0), productBond=4, status='New'), BondInfo(product=0, productAtoms=(2, 1), productBond=1, status='Changed'), BondInfo(product=0, productAtoms=(3, 2), productBond=2, status='New'), BondInfo(product=0, productAtoms=(4, 3), productBond=3, status='Changed')]Finally, define the funciton which we’ll use to draw the product molecule with highlights shown for bonds and atoms involved in the reaction:
from IPython.display import Image
def draw_product_with_modified_bonds(rxn,atms,bnds,productIdx=None,showAtomMaps=False):
if productIdx is None:
pcnts = [x.GetNumAtoms() for x in rxn.GetProducts()]
largestProduct = list(sorted(zip(pcnts,range(len(pcnts))),reverse=True))[0][1]
productIdx = largestProduct
d2d = Draw.rdMolDraw2D.MolDraw2DCairo(350,300)
pmol = Chem.Mol(rxn.GetProductTemplate(productIdx))
Chem.SanitizeMol(pmol)
if not showAtomMaps:
for atom in pmol.GetAtoms():
atom.SetAtomMapNum(0)
bonds_to_highlight=[]
highlight_bond_colors={}
atoms_seen = set()
for binfo in bnds:
if binfo.product==productIdx and binfo.status=='New':
bonds_to_highlight.append(binfo.productBond)
atoms_seen.update(binfo.productAtoms)
highlight_bond_colors[binfo.productBond] = (1,.4,.4)
if binfo.product==productIdx and binfo.status=='Changed':
bonds_to_highlight.append(binfo.productBond)
atoms_seen.update(binfo.productAtoms)
highlight_bond_colors[binfo.productBond] = (.4,.4,1)
atoms_to_highlight=set()
for ainfo in atms:
if ainfo.product != productIdx or ainfo.productAtom in atoms_seen:
continue
atoms_to_highlight.add(ainfo.productAtom)
d2d.drawOptions().useBWAtomPalette()
d2d.drawOptions().continuousHighlight=False
d2d.drawOptions().highlightBondWidthMultiplier = 24
d2d.drawOptions().setHighlightColour((.9,.9,0))
d2d.drawOptions().fillHighlights=False
atoms_to_highlight.update(atoms_seen)
d2d.DrawMolecule(pmol,highlightAtoms=atoms_to_highlight,highlightBonds=bonds_to_highlight,
highlightBondColors=highlight_bond_colors)
d2d.FinishDrawing()
return d2d.GetDrawingText()Now we can draw the highlighted product molecule:
Let’s look at some other reactions. I will use the SI data from
| rxn_Class | patentID | rxnSmiles_Mapping_NameRxn | reactantSet_NameRxn | NameRxn_Mapping_Complete | rxnSmiles_Mapping_IndigoTK | reactantSet_IndigoTK | IndigoTK_Mapping_Complete | rxnSmiles_IndigoAutoMapperKNIME | reactantSet_IndigoAutoMapperKNIME | IndigoAutoMapperKNIME_Mapping_Complete | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | US05849732 | C.CCCCCC.CO.O=C(OCc1ccccc1)[NH:1][CH2:2][CH2:3... | set([3, 4]) | True | C(OC([NH:11][CH2:12][CH2:13][CH2:14][CH2:15][C... | set([0, 2]) | True | C.CCCCCC.CO.[CH3:10][O:11][C:12]([C@@H:14]([NH... | set([3, 4]) | True |
| 1 | 2 | US20120114765A1 | O[C:1](=[O:2])[c:3]1[cH:4][c:5]([N+:6](=[O:7])... | set([0, 1]) | True | [Cl:1][c:2]1[cH:3][n:4][cH:5][c:6]([Cl:20])[c:... | set([0, 1]) | True | [NH2:1][c:2]1[c:11]2[c:6]([cH:7][n:8][cH:9][cH... | set([0, 1]) | True |
| 2 | 1 | US08003648B2 | Cl.O=[CH:1][c:2]1[cH:3][cH:4][c:5](-[c:6]2[n:7... | set([1, 3]) | True | [CH2:1]([NH:3][CH2:4][CH3:5])[CH3:2].C([BH3-])... | set([0, 3]) | True | [CH3:1][CH2:2][NH:3][CH2:4][CH3:5].[CH3:6][c:7... | set([0, 1]) | True |
| 3 | 1 | US09045475B2 | CC(=O)O[BH-](OC(C)=O)OC(C)=O.ClCCl.O=[C:1]([CH... | set([2, 3]) | True | [nH:1]1[c:5]2[n:6][cH:7][c:8]([O:10][c:11]3[cH... | set([0, 3]) | True | CC(O[BH-](OC(=O)C)OC(=O)C)=O.[CH3:14][C:15]1([... | set([1, 3]) | True |
| 4 | 2 | US08188098B2 | CCN(C(C)C)C(C)C.ClCCl.Cl[C:1](=[O:2])[O:3][CH:... | set([2, 5]) | True | Cl[C:2]([O:4][CH:5]1[CH2:9][CH2:8][CH2:7][CH2:... | set([0, 2]) | True | CCN(C(C)C)C(C)C.[CH3:10][CH2:11][O:12][c:13]1[... | set([1, 4]) | True |
rxnclass = 1
class_smis = df[df['rxn_Class']==rxnclass].rxnSmiles_Mapping_NameRxn.to_list()
rxn = rdChemReactions.ReactionFromSmarts(class_smis[3],useSmiles=True)
rxn.Initialize()
atms,bnds = find_modifications_in_products(rxn)
print(atms)
print(bnds)
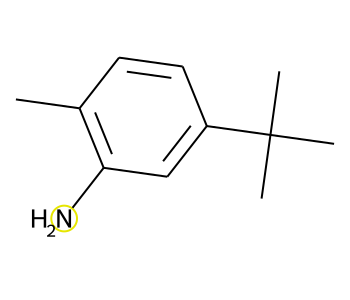
Image(draw_product_with_modified_bonds(rxn,atms,bnds))[AtomInfo(mapnum=1, reactant=3, reactantAtom=1, product=0, productAtom=0), AtomInfo(mapnum=7, reactant=4, reactantAtom=0, product=0, productAtom=6)]
[BondInfo(product=0, productAtoms=(6, 0), productBond=5, status='New')]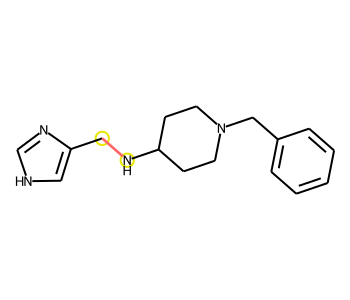
rxnclass = 4
class_smis = df[df['rxn_Class']==rxnclass].rxnSmiles_Mapping_NameRxn.to_list()
rxn = rdChemReactions.ReactionFromSmarts(class_smis[1],useSmiles=True)
rxn.Initialize()
atms,bnds = find_modifications_in_products(rxn)
print(atms)
print(bnds)
Image(draw_product_with_modified_bonds(rxn,atms,bnds))[AtomInfo(mapnum=1, reactant=1, reactantAtom=1, product=0, productAtom=0), AtomInfo(mapnum=2, reactant=1, reactantAtom=2, product=0, productAtom=9), AtomInfo(mapnum=16, reactant=2, reactantAtom=0, product=0, productAtom=18), AtomInfo(mapnum=17, reactant=2, reactantAtom=1, product=0, productAtom=16), AtomInfo(mapnum=19, reactant=2, reactantAtom=3, product=0, productAtom=15)]
[BondInfo(product=0, productAtoms=(9, 0), productBond=8, status='Changed'), BondInfo(product=0, productAtoms=(18, 0), productBond=18, status='New'), BondInfo(product=0, productAtoms=(15, 9), productBond=14, status='New'), BondInfo(product=0, productAtoms=(16, 18), productBond=17, status='Changed'), BondInfo(product=0, productAtoms=(15, 16), productBond=15, status='Changed')]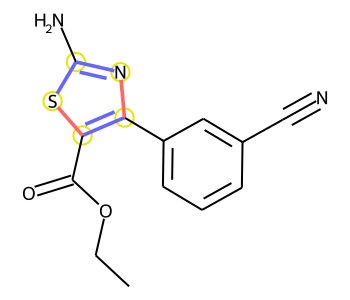
Look at an example where there are no changed bonds in the products but where there is a changed atom:
rxnclass = 7
class_smis = df[df['rxn_Class']==rxnclass].rxnSmiles_Mapping_NameRxn.to_list()
rxn = rdChemReactions.ReactionFromSmarts(class_smis[1],useSmiles=True)
rxn.Initialize()
atms,bnds = find_modifications_in_products(rxn)
print(atms)
print(bnds)
Image(draw_product_with_modified_bonds(rxn,atms,bnds))[AtomInfo(mapnum=1, reactant=1, reactantAtom=1, product=0, productAtom=0)]
[]